Education Research

Part of my research team investigates how students learn genomics in authentic research experiences using Chamaecrista transcriptomes. Authentic research experiences provide an opportunity to scale the benefits of undergraduate research to the level of whole courses. More information is available on the Education page of this site.
When Partridge Peas (Chamaecrista fasciculata ) Flower – Gene x Enviroment Interactions


With Jeff Doyle (Cornell), Marc Libault (University of Oklahoma) and Steven Cannon (Iowa State), we are developing Chamaecrista fasciculata as a model organism to test key questions about the evolution of the Leguminosae (a large family of plants bearing bean pods), several members of which are important agricultural crops or key model organisms for genetic analysis. These questions include:
1) whether polyploidy is an ancient feature of this family, or characteristic of the more recently-evolved species only (Cannon. S.B., D. Illut, A.D, Farmer, S.L. Maki, G.D. May, S.R. Singer, J.J. Doyle (2010) Nodule evolution did not depend on early polyploidy in the legumes. PLoS ONE, 5(7): e11630. doi:10.1371/journal.pone.0011630),
2) whether nodules containing symbiotic bacteria in C. fasciculata evolved independently from those common in other legumes (Young, N.D. et al. (2011) The Medicago genome provides insight into the evolution of rhizobial symbioses. Nature. doi:10.1038/nature10625), and
3) which genes for flowering time and flower structure are unique to the legumes, conserved within the legumes, and divergent in the more recent legumes.
We are using RNA-Seq analysis to compare expression levels in ecotypes grown under different temperature and light regimes. To sort out environmental effects, we are studying annual variations in growth and flowering time of C. fasciculata in three native prairie sites in Minnesota.

For more detail, see our November 2009 Plant Physiology article.
Genetic Regulation of Inflorescence Architecture in Pea
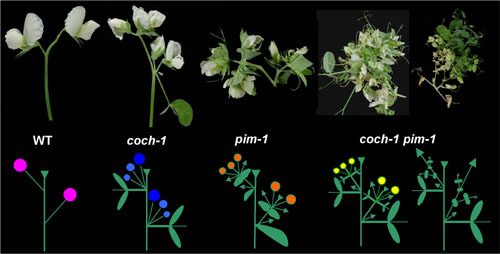
The evolution of inflorescence architecture involves a common toolkit of flowering genes used in different ways to create diverse morphologies, which can dramatically affect reproductive success. Using a candidate gene approach, we are identifying genes that correspond to known inflorescence phenotypes in pea. We are interested in the interactions among these genes in terms of phenotype and gene expression. We are particularly interested in COCH, PIM, DET, UNI, and VEG1 interact to regulate determinancy in inflorescences. Below are images of wild type, coch, pim, and coch pim plants and a few sketches of their branching patterns of the axillary inflorescence.
Modeling Genetic Regulation of Inflorescence Development
Creating and comparing systems-based models of shoot development in different species may reveal subtle variations that result in distinct patterns. To test hypotheses about gene interaction and aid in the design of future experiments, we are using L-systems to create process-based models of inflorescence development. This work is in collaboration with our colleagues Jim Hanan and Christine Beveridge at the University of Queensland. We do our modeling work using L-Studio.